摘要: Separating densely packed molecules before imaging allows them to become visible for the first time.
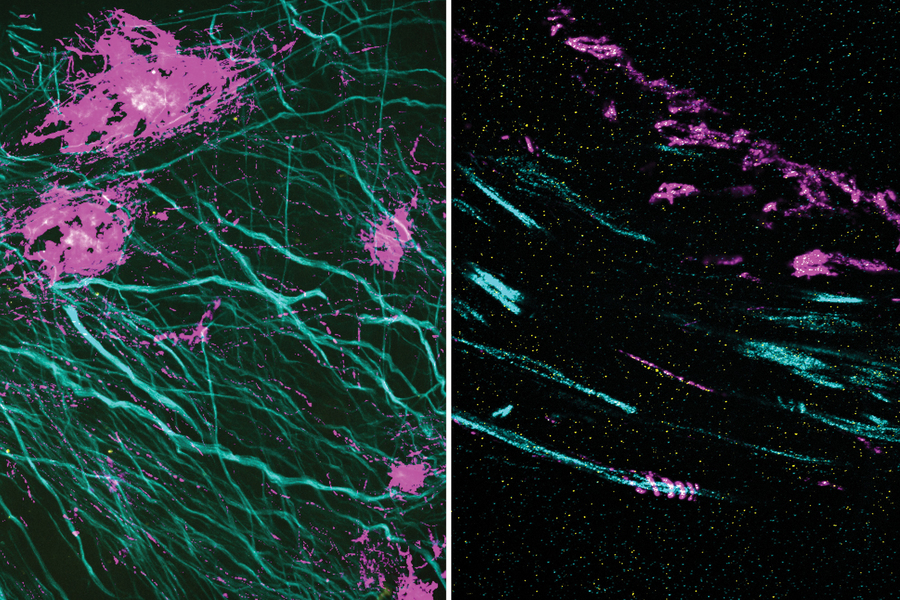
▲圖片來源:mit
Inside a living cell, proteins and other molecules are often tightly packed together. These dense clusters can be difficult to image because the fluorescent labels used to make them visible can’t wedge themselves in between the molecules.
MIT researchers have now developed a novel way to overcome this limitation and make those “invisible” molecules visible. Their technique allows them to “de-crowd” the molecules by expanding a cell or tissue sample before labeling the molecules, which makes the molecules more accessible to fluorescent tags.
This method, which builds on a widely used technique known as expansion microscopy previously developed at MIT, should allow scientists to visualize molecules and cellular structures that have never been seen before.
“It’s becoming clear that the expansion process will reveal many new biological discoveries. If biologists and clinicians have been studying a protein in the brain or another biological specimen, and they’re labeling it the regular way, they might be missing entire categories of phenomena,” says Edward Boyden, the Y. Eva Tan Professor in Neurotechnology, a professor of biological engineering and brain and cognitive sciences at MIT, a Howard Hughes Medical Institute investigator, and a member of MIT’s McGovern Institute for Brain Research and Koch Institute for Integrative Cancer Research.
Using this technique, Boyden and his colleagues showed that they could image a nanostructure found in the synapses of neurons. They also imaged the structure of Alzheimer’s-linked amyloid beta plaques in greater detail than has been possible before.
“Our technology, which we named expansion revealing, enables visualization of these nanostructures, which previously remained hidden, using hardware easily available in academic labs,” says Deblina Sarkar, an assistant professor in the Media Lab and one of the lead authors of the study.
The senior authors of the study are Boyden; Li-Huei Tsai, director of MIT’s Picower Institute for Learning and Memory; and Thomas Blanpied, a professor of physiology at the University of Maryland. Other lead authors include Jinyoung Kang, an MIT postdoc, and Asmamaw Wassie, a recent MIT PhD recipient. The study appears today in Nature Biomedical Engineering.
De-crowding
Imaging a specific protein or other molecule inside a cell requires labeling it with a fluorescent tag carried by an antibody that binds to the target. Antibodies are about 10 nanometers long, while typical cellular proteins are usually about 2 to 5 nanometers in diameter, so if the target proteins are too densely packed, the antibodies can’t get to them.
This has been an obstacle to traditional imaging and also to the original version of expansion microscopy, which Boyden first developed in 2015. In the original version of expansion microscopy, researchers attached fluorescent labels to molecules of interest before they expanded the tissue. The labeling was done first, in part because the researchers had to use an enzyme to chop up proteins in the sample so the tissue could be expanded. This meant that the proteins couldn’t be labeled after the tissue was expanded.
To overcome that obstacle, the researchers had to find a way to expand the tissue while leaving the proteins intact. They used heat instead of enzymes to soften the tissue, allowing the tissue to expand 20-fold without being destroyed. Then, the separated proteins could be labeled with fluorescent tags after expansion.
With so many more proteins accessible for labeling, the researchers were able to identify tiny cellular structures within synapses, the connections between neurons that are densely packed with proteins. They labeled and imaged seven different synaptic proteins, which allowed them to visualize, in detail, “nanocolumns” consisting of calcium channels aligned with other synaptic proteins. These nanocolumns, which are believed to help make synaptic communication more efficient, were first discovered by Blanpied’s lab in 2016.
“This technology can be used to answer a lot of biological questions about dysfunction in synaptic proteins, which are involved in neurodegenerative diseases,” Kang says. “Until now there has been no tool to visualize synapses very well.”
New patterns
The researchers also used their new technique to image beta amyloid, a peptide that forms plaques in the brains of Alzheimer’s patients. Using brain tissue from mice, the researchers found that amyloid beta forms periodic nanoclusters, which had not been seen before. These clusters of amyloid beta also include potassium channels. The researchers also found amyloid beta molecules that formed helical structures along axons.
“In this paper, we don’t speculate as to what that biology might mean, but we show that it exists. That is just one example of the new patterns that we can see,” says Margaret Schroeder, an MIT graduate student who is also an author of the paper.
Sarkar says that she is fascinated by the nanoscale biomolecular patterns that this technology unveils. “With a background in nanoelectronics, I have developed electronic chips that require extremely precise alignment, in the nanofab. But when I see that in our brain Mother Nature has arranged biomolecules with such nanoscale precision, that really blows my mind,” she says.
Boyden and his group members are now working with other labs to study cellular structures such as protein aggregates linked to Parkinson’s and other diseases. In other projects, they are studying pathogens that infect cells and molecules that are involved in aging in the brain. Preliminary results from these studies have also revealed novel structures, Boyden says.
“Time and time again, you see things that are truly shocking,” he says. “It shows us how much we are missing with classical unexpanded staining.”
The researchers are also working on modifying the technique so they can image up to 20 proteins at a time. They are also working on adapting their process so that it can be used on human tissue samples.
Sarkar and her team, on the other hand, are developing tiny wirelessly powered nanoelectronic devices which could be distributed in the brain. They plan to integrate these devices with expansion revealing. “This can combine the intelligence of nanoelectronics with the nanoscopy prowess of expansion technology, for an integrated functional and structural understanding of the brain,” Sarkar says.
The research was funded by the National Institutes of Health, the National Science Foundation, the Ludwig Family Foundation, the JPB Foundation, the Open Philanthropy Project, John Doerr, Lisa Yang and the Tan-Yang Center for Autism Research at MIT, the U.S. Army Research Office, Charles Hieken, Tom Stocky, Kathleen Octavio, Lore McGovern, Good Ventures, and HHMI.
轉貼自: mit.edu
若喜歡本文,請關注我們的臉書 Please Like our Facebook Page: Big Data In Finance

留下你的回應
以訪客張貼回應